t-SNE (t-distributed Stochastic Neighbor Embedding) is a popular method for exploring high-dimensional data proposed by Hinton and van der Maaten in 2008. The technique has become widespread in the machine learning community, mostly because of its magical ability to create compelling two-dimensional “visualization” from very high-dimensional data.
t-SNE as a dimensionality reduction method attempts to preserve most of the local structure of the data by matching similarity distributions. Even though the method has proven to yield very interesting 2D “maps” of the high-dimensional data, current implementations struggle to perform this visualization on large-scale datasets.
For this reason, researchers from the University of California, Berkeley, have proposed a CUDA implementation of t-SNE, which offers enormous speedups in terms of processing time. According to them, their GPU-accelerated t-SNE outperforms existing implementations with 50-700x speedups on visualizing CIFAR-10 and MNIST datasets.
Researchers demonstrate the power of the proposed t-SNE CUDA by visualizing previously computationally intractable visualizations. For example, they visualized the neural network activations on the entire ImageNet dataset in the Computer Vision domain and all of the GloVe embedding vectors in the NLP domain. In their paper, they mention that t-SNE CUDA will enable visualizations of a large number of things that previously were intractable
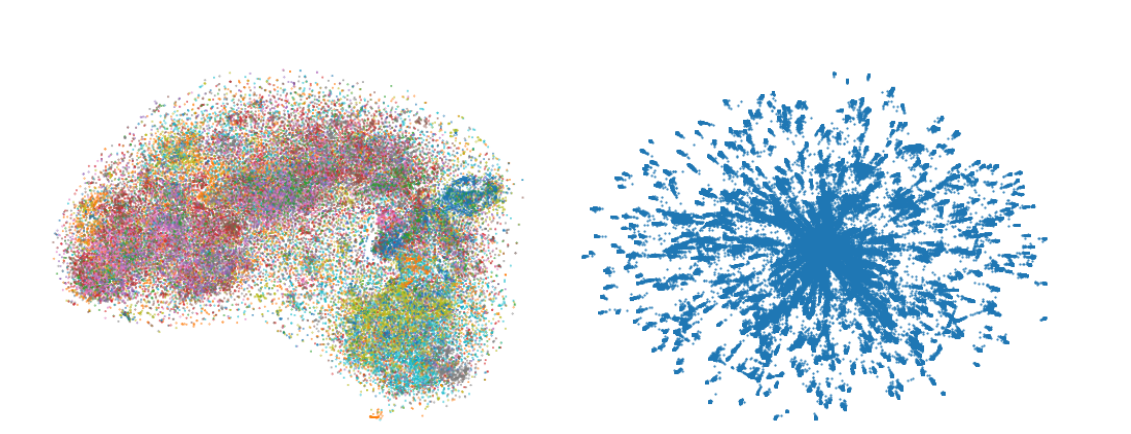
using t-SNE-CUDA. [Right] Embedding of the 1.2M VGG16 Codes (4096 dim)
computed in 523s.