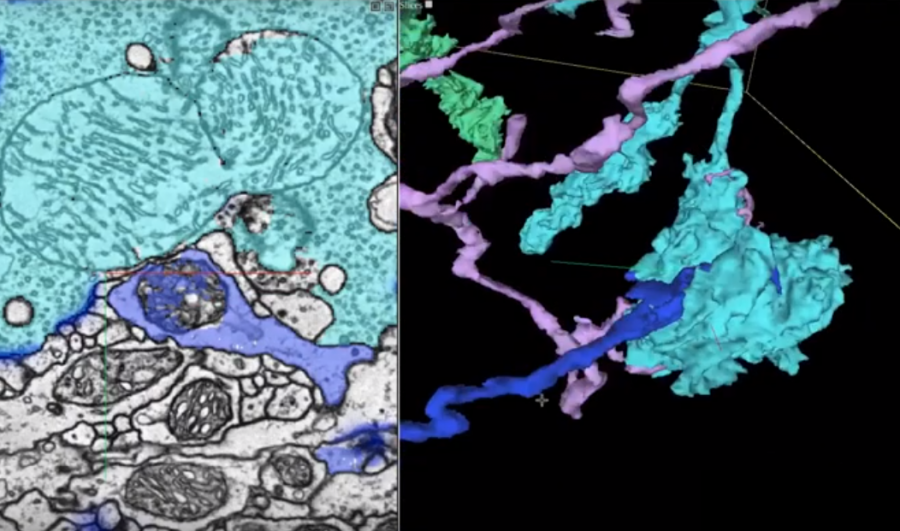
Researchers from Google, in collaboration with researchers from Janelia Research Campus and Cambridge University, have presented a method for fully automated reconstruction of an entire fruit fly brain.
In their work, the researchers combined several advances from different fields such as biology, computer graphics, and machine learning in order to map the whole “wiring diagram” of the brain. A transmission electron microscope was used to image each of the thousands of ultra-thin slices of the sectioned fruit fly brain. These images were aligned into a coherent 3D volume of the brain and machine learning was used to trace each individual neuron of the brain.
Flood-filling networks – a special type of convolutional neural networks for segmentation was used to find the traces of the neurons by segmenting each of the images in the obtained 3D volume. These networks are able to find the trace of each neuron due to their recurrent pathways that allow iterative optimization. Researchers added two new procedures to improve the results obtained from the FFN models. In fact, they first estimated the slice-to-slice consistency in the 3D volume to stabilize the content and they used a generative model – SECGAN (Segmentation-Enhanced Cycle GAN) to fill the missing slices in the volume. Researchers mention that the flood-filling network was able to find the traces much better when using SECGAN augmented data as the base.

Together with this announcement, the researchers announced the release of a new tool for visualizing 3D images, which is specifically designed for visualizing neuron connections. The powerful tool named Neuroglancer helped the researchers during their work on this project and was released as an open-source project.
More details about the project can be found in the blog post or in the paper published in Biorxiv. The visualization tool is available on Github.