Medical image segmentation plays a critical role in precision medicine, enabling more accurate diagnosis, treatment planning, and quantitative analysis. While significant progress has been made in developing both specialized and generalist segmentation models for 2D medical images, the landscape of 3D and video segmentation remains underexplored. A new breakthrough, MedSAM2, addresses this gap by providing a foundation model for 3D medical images and videos, built upon the Segment Anything Model 2 (SAM2) architecture. Most importantly, MedSAM2 is completely open-source, with code, model weights, and annotated datasets publicly available on Github.
Core Technical Innovation
MedSAM2 represents a significant advancement in medical imaging AI through several key innovations:
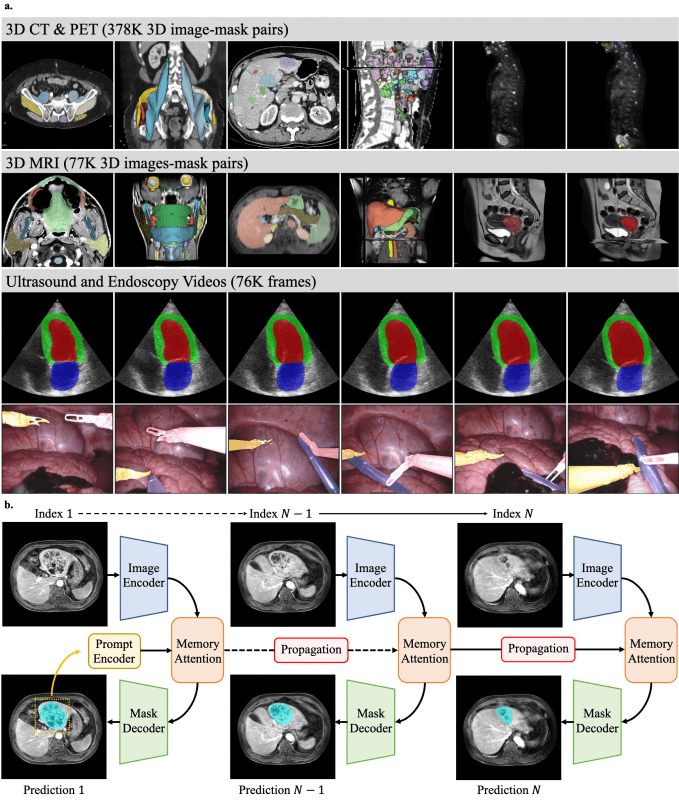
- Architecture Optimization: Built on SAM2’s foundation with modifications for medical domain specificity, particularly for 3D data processing. The model utilizes the hierarchical vision transformer (Hiera) backbone with a memory attention module that efficiently handles spatial continuity across volumetric slices or video frames.
- Comprehensive Training Dataset: Fine-tuned on an extensive dataset containing over 455,000 3D image-mask pairs spanning CT (363,161), PET (14,818), and MRI (77,154), plus 76,000 video frames from ultrasound (19,232) and endoscopy (56,462).
- Memory-Conditioned Processing: Employs a streaming memory bank that conditions current frame features on previous frames’ predictions, enabling effective propagation of segmentation masks across 3D volumes or sequential video frames.
- Transfer Learning Approach: Achieves superior performance through full-model fine-tuning of the lightweight SAM2.1-Tiny variant, with differential learning rates – lower for the image encoder to preserve pre-trained feature extraction capabilities and higher for other components to adapt to medical domain characteristics.
Performance Metrics
MedSAM2 demonstrates significant improvements over existing segmentation models in comprehensive evaluations. Below are comparative performance metrics showing MedSAM2 against SAM2.1 variants (Tiny, Small, Base, Large) and EfficientMedSAM-Top1:
3D Image Segmentation (Dice Similarity Coefficient)
| Task | MedSAM2 | EfficientMedSAM-Top1 | Best SAM2.1 Variant |
|---|---|---|---|
| CT organs | 88.84% (80.03-94.03%) | 83.55% (67.20-91.78%) | ~80% (various models) |
| CT lesions | 86.68% (74.32-91.14%) | 77.95% (69.15-84.81%) | ~70% (various models) |
| MRI organs | 87.06% (82.96-90.04%) | 74.83% | ~84% (SAM2.1-Base) |
| MRI lesions | 88.37% (79.91-93.26%) | 82.25% (68.30-90.53%) | ~72% (various models) |
| PET lesions | 87.22% (79.07-90.45%) | 77.85% | ~80% (SAM2.1-Large) |
Key observations:
- MedSAM2 outperforms EfficientMedSAM-Top1 by 5-13% across different tasks
- All SAM2.1 variants performed similarly regardless of model size (no statistically significant differences, p>0.05)
- The performance gap is particularly pronounced for lesions, which are traditionally more challenging to segment
Video Segmentation (Dice Similarity Coefficient)
| Task | MedSAM2 | SAM2.1-Tiny | SAM2.1-Small | SAM2.1-Base | SAM2.1-Large |
|---|---|---|---|---|---|
| Ultrasound LV | 96.13% (95.09-97.15%) | ~94% | ~94% | ~94% | ~94% |
| Ultrasound LV epicardium | 93.10% (91.07-94.11%) | ~88% | ~89% | ~89% | ~90% |
| Ultrasound LA | 95.79% (94.38-96.96%) | ~93% | ~93% | ~94% | ~94% |
| Endoscopy polyps (easy) | 92.24% (85.15-96.11%) | 92.11% (75.74-96.47%) | 93.32% (76.24-96.58%) | 93.87% (77.48-96.64%) | 93.76% (77.20-96.60%) |
| Endoscopy polyps (hard) | 92.22% (83.37-95.88%) | 83.43% (60.34-92.53%) | 84.93% (63.32-92.87%) | 85.64% (64.55-92.98%) | 87.47% (67.21-93.51%) |
Notable findings:
- For cardiac ultrasound, MedSAM2 shows 2-4% improvement over SAM2.1 variants
- For hard polyp cases, MedSAM2 maintains consistent performance with a 5-9% improvement over SAM2.1 models
- MedSAM2 exhibits significantly reduced variability (narrower interquartile ranges) across all tasks, indicating more robust and reliable segmentation
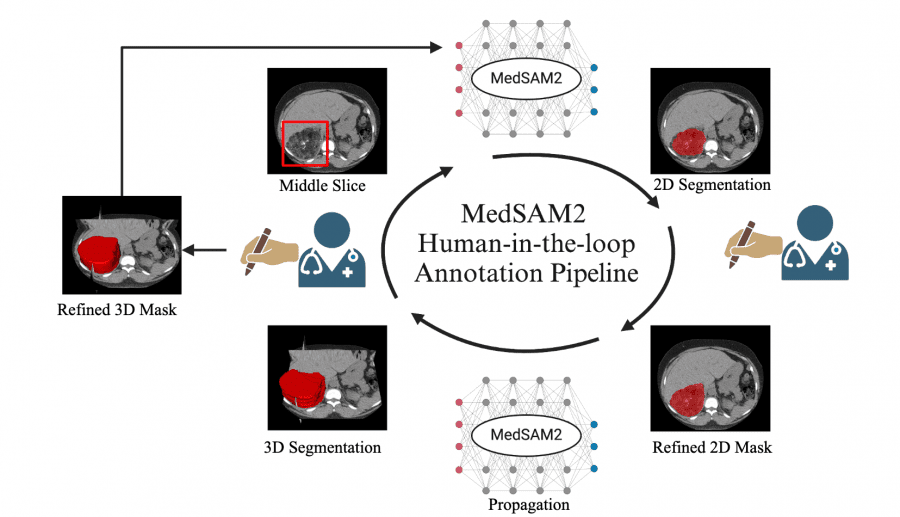
Human-in-the-Loop Annotation Pipeline
Perhaps the most significant contribution of MedSAM2 is its practical application in data annotation workflows. The researchers implemented a human-in-the-loop annotation pipeline that dramatically reduces manual annotation time:
- CT Lesion Annotation: Reduced annotation time from 525.9 seconds to 74.3 seconds per lesion (85.86% faster) over three iterative rounds, enabling the annotation of 5,000 CT lesions.
- MRI Liver Lesion Annotation: Decreased annotation time from 520.3 seconds to 65.2 seconds per lesion (87.47% faster), facilitating the annotation of 3,984 liver MRI lesions.
- Echocardiography Video Annotation: Cut annotation time from 102.3 seconds to 8.4 seconds per frame (91.79% faster), allowing for the annotation of 251,550 frames across 1,800 videos.
This iterative process demonstrates how model performance improves with each round of human feedback, creating a virtuous cycle that continuously enhances annotation efficiency.
Open-Source Availability and Deployment
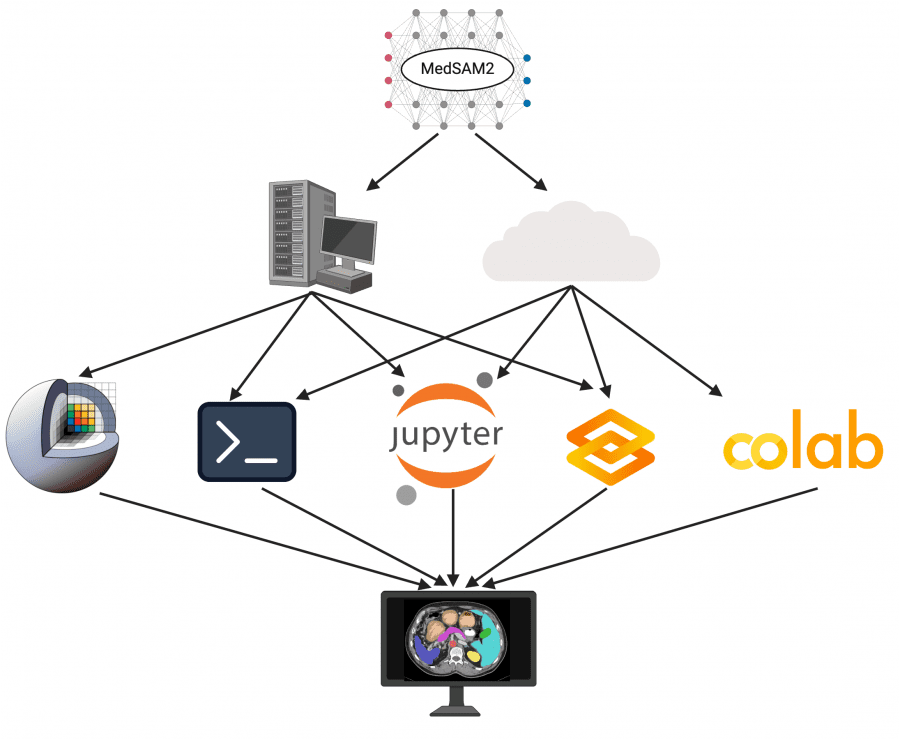
A key strength of MedSAM2 is its fully open-source nature, providing the research and clinical communities with complete access to:
- Complete codebase: All implementation details and training pipelines
- Pre-trained model weights: Ready-to-use models without requiring expensive retraining
- Annotated datasets: Valuable resources for benchmarking and further research
- 3D Slicer plugin: Open-source extension for the widely-used medical imaging platform
The researchers implemented MedSAM2 across multiple platforms to enhance accessibility:
- 3D Slicer Plugin: Integration with this widely-used open-source medical imaging platform enables seamless clinical and research application (available at https://github.com/bowang-lab/MedSAMSlicer).
- Command-line Interface: For high-throughput batch processing of large datasets.
- JupyterLab and Colab: For interactive, code-centric experimentation and development.
- Gradio: A lightweight web-based interface for users without extensive technical expertise or computational resources.
This open-source approach facilitates community collaboration, enables customization for specific clinical workflows, and democratizes access to advanced segmentation technology regardless of institutional resources.
Technical Limitations
Despite its impressive capabilities, MedSAM2 has several limitations:
- Bounding Box Dependency: Reliance on bounding boxes as primary prompts limits segmentation capabilities for complex anatomical structures like thin, branching vessels.
- Fixed Memory Design: The eight-frame memory bank may be insufficient for rapid or large object movements, potentially causing tracking failures.
- Computational Requirements: Despite using the lightweight SAM2.1-Tiny variant, inference still requires GPU computation, limiting deployment in resource-constrained environments.
Future Directions
Potential advancements include:
- 4D Image Encoder: Jointly processing spatial and temporal information for better contextualization.
- Alternative Prompting Methods: Supporting point, text, scribble, and lasso prompts for more flexible interaction.
- Adaptive Memory System: Implementing variable memory retention for different segmentation targets.
- Model Optimization: Further compression, quantization, and distillation to enable efficient CPU-based inference.
Conclusion
MedSAM2 represents a significant advancement in medical image analysis, bridging the gap between general foundation models and domain-specific medical applications. Its ability to handle both volumetric scans and video sequences, coupled with substantial annotation efficiency improvements, makes it a valuable tool for both research and clinical deployment. By dramatically reducing the annotation burden, MedSAM2 facilitates the creation of larger, higher-quality datasets that will further drive progress in medical AI.
The open-source nature of this project cannot be overstated in its importance. By providing free access to code, model weights, annotated datasets, and deployment tools, the researchers have eliminated significant barriers to entry in advanced medical image segmentation. This approach enables:
- Replication and validation of results by independent researchers
- Customization for specific clinical needs and imaging protocols
- Continuous improvement through community contributions
- Educational applications for training medical imaging specialists
- Democratized access across resource-constrained healthcare settings globally
The comprehensive open-source release, combined with integration across established platforms, will likely accelerate adoption and community-driven improvements, potentially transforming workflows across cardiology, oncology, and surgical specialties where precise 3D segmentation is critical but traditionally time-consuming.