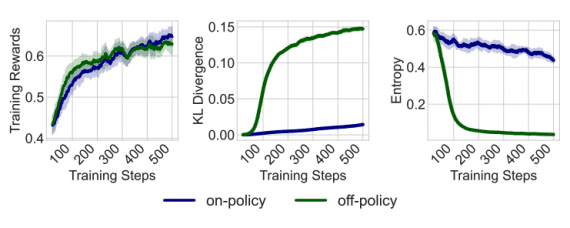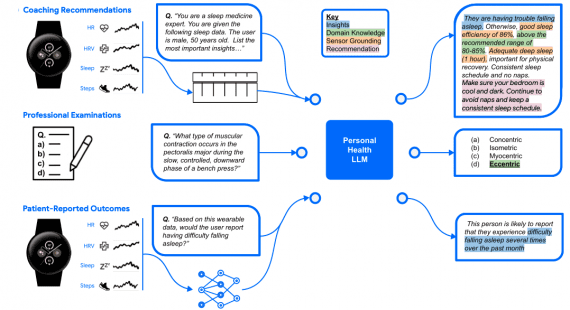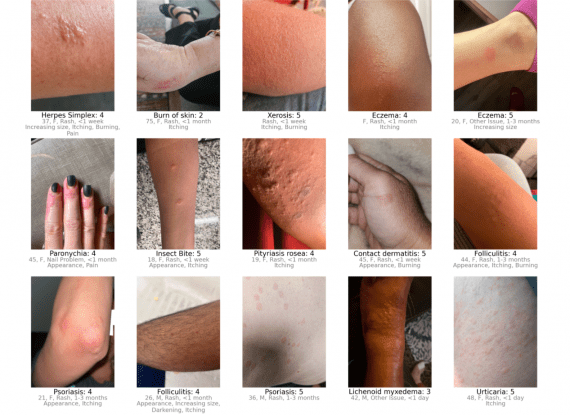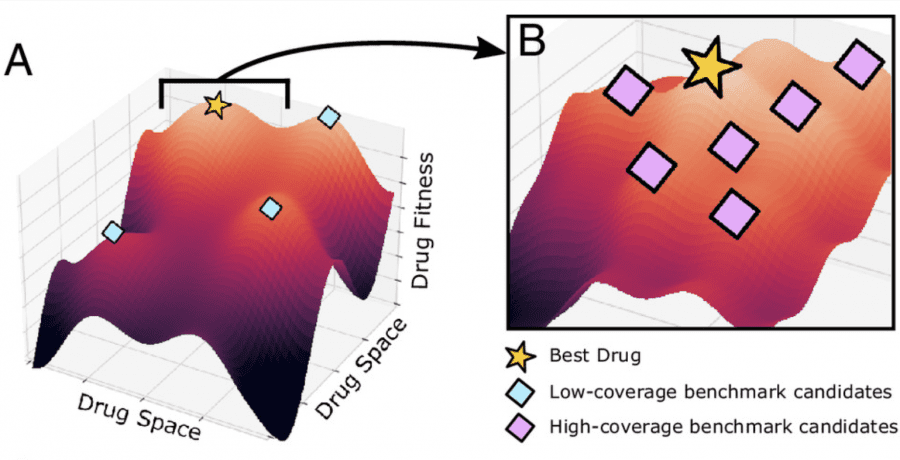
ConPLex is a language model trained to analyze chemical databases and search for potential drug molecules that interact best with specific target proteins. The model enables the exploration of over 100 million compounds per day, significantly speeding up the search for new medications.
Huge databases of chemical compounds can hold potential treatment methods for various diseases. In recent years, scientists have started utilizing computational methods to scan these databases in hopes of accelerating drug discovery. However, many of these methods require extensive time because most of them compute the three-dimensional structure of each protein based on its amino acid sequence.
ConPLex can match target proteins with potential drug molecules without the need for structure calculation. Working with a database of over 20,000 proteins, the model encodes their data into numerical representations of each amino acid sequence, capturing the relationship between sequence and structure. Proteins with dissimilar sequences but potentially similar structures can be represented similarly in the language space.
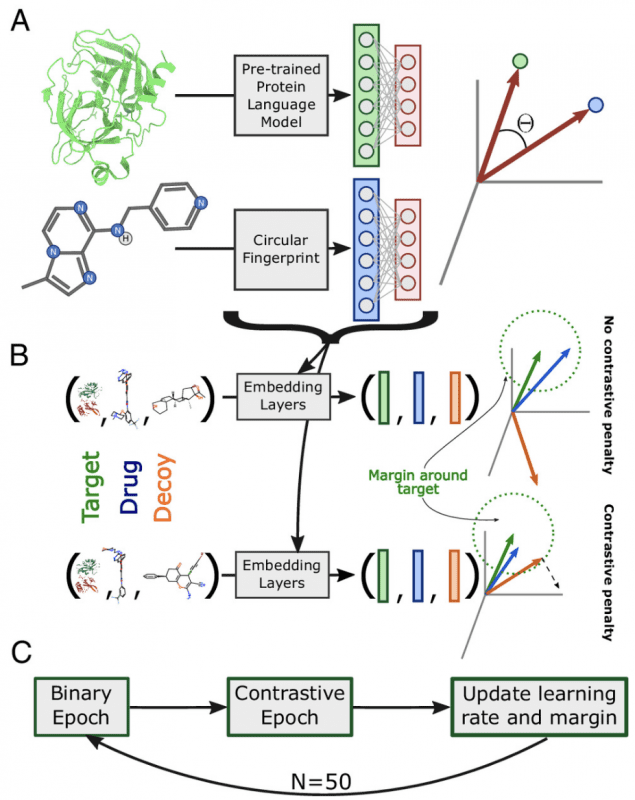
ConPLex was trained on existing medications. Researchers tested the model by scanning a library of approximately 4,700 candidate molecules for their ability to bind to a set of 51 enzymes and then experimentally verifying its predictions. Out of the 19 molecule-protein pairs predicted by the model, 12 exhibited strong binding affinity. Four of these pairs showed extremely high affinity, similar to that typically achieved by medicinal drugs.
Scientists plan to expand the model to high-molecular-weight compounds. This type of modeling could also be useful for screening the toxicity of potential drug compounds before animal testing.